For Transmission Recording

Stimulated Emission Depletion (STED) is a powerful super resolution microscopy technique that allows for the observation of fluorescence structures with spatial resolution below the diffraction limit. The Alba STED uses the pulsed excitation and pulsed depletion approach (pSTED) to achieve lateral resolution of 60 - 100 nm. An additional increase on the resolution is provided by the lifetime information acquired through FastFLIM (the digital frequency domain lifetime imaging) and using the phasor plot: the photons emitted by the fluorophores activated by the excitation laser only (inherent fluorescence decay) are efficiently separated from the photons emitted by the fluorophores partially or fully activated by the STED laser (featuring shorter decay times). A resolution of 30 nm is measured on standard samples. Moreover, the combination of pSTED and lifetime analysis through the phasor plot allows to separate two fluorophores featuring similar emission patterns but having different decay times (dual label excitation).

High Dynamic Range

Fast Image Acquisition
(Dwell Time: 0.2 µs)

pSTED
(Pulsed Excitation and Pulsed STED)
w/ FastFLIM for Time-Resolved Acquisition

Improved Image Resolution
Using Lifetime Information
and the Phasor Plot

Dual Label Excitation


A peristaltic pump supplies the stage with a solution for keeping the sample conditions (temperature, pH, etc...) stable.

When using water objectives for prolonged measurements, it prevents the liquid drying up.

It keeps the focus position of the objective for hours using an active feedback to counter drifts.

Stage top incubator or a full enclosure to maintain the environmental conditions of cell cultures.

Visualize your sample with the Epi module. Select as light source either an arc lamp or an LED and the suitable filter cubes to add to the microscope cassette.

Fully integrated for the models:
NanoWizard by JPK-Bruker
Resolve by Bruker
For other models contact ISS.
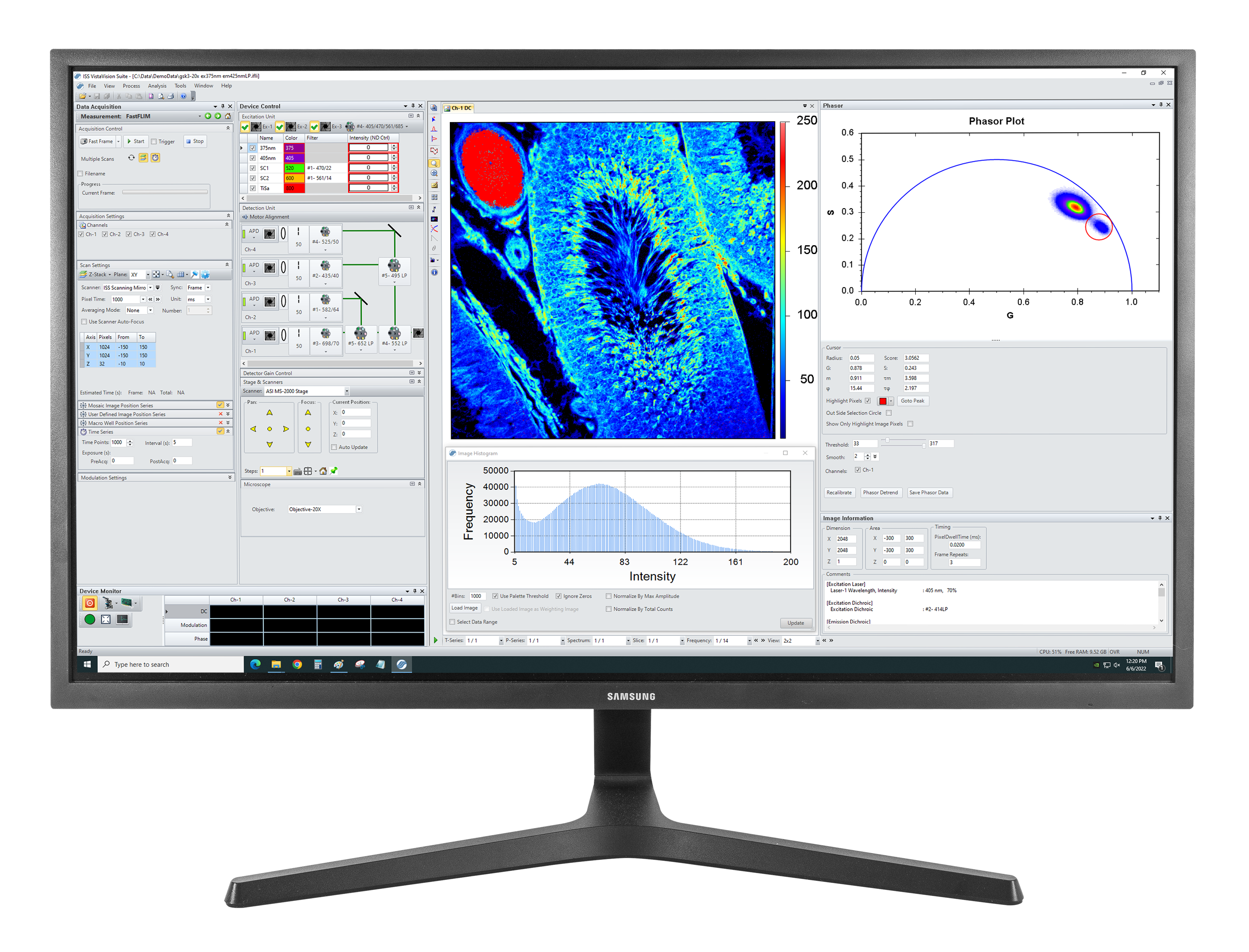
VistaVision is a complete software package for confocal microscopy applications including instrument control, data acquisition and data processing. Easy to use, the software has been developed in modular components that can be activated when a specific instrument configuration is selected. The modules include: